About
Biography
Hola! My name is Erick Armingol, and I am currently a Postdoctoral Fellow at the Wellcome Sanger Institute. I work in the Vento Lab, wherein my research focus is on placental development, in both eutopic and ectopic pregnancy, and the effect of menopause on homeostasis and the dynamic resilience of women. By combining machine learning and state-of-the-art tools, I aim to find gene expression changes and elucidate how they impact cellular functions by inspecting their metabolism and cell-cell interactions.
I did my PhD research in the Lewis Lab at the University of California, San Diego. There, I focused on developing methods for inferring cell-cell interactions from single-cell data.
My research interests are in using single-cell technologies for inferring cell-cell interactions, modeling metabolic fluxes, and analyzing signaling pathways.
Education
PhD in Bioinformatics and Systems Biology
University of California, San Diego (2018 - 2023)
Engineer in Molecular Biotechnology
Universidad de Chile (2009 - 2015)
BS in Molecular Biotechnology
Universidad de Chile (2009 - 2013)
Interests
- Bioinformatics
- Systems Biology
- Biological Modeling
- Machine Learning
- Tool Development
You can find me in
Cell-cell interactions and communication
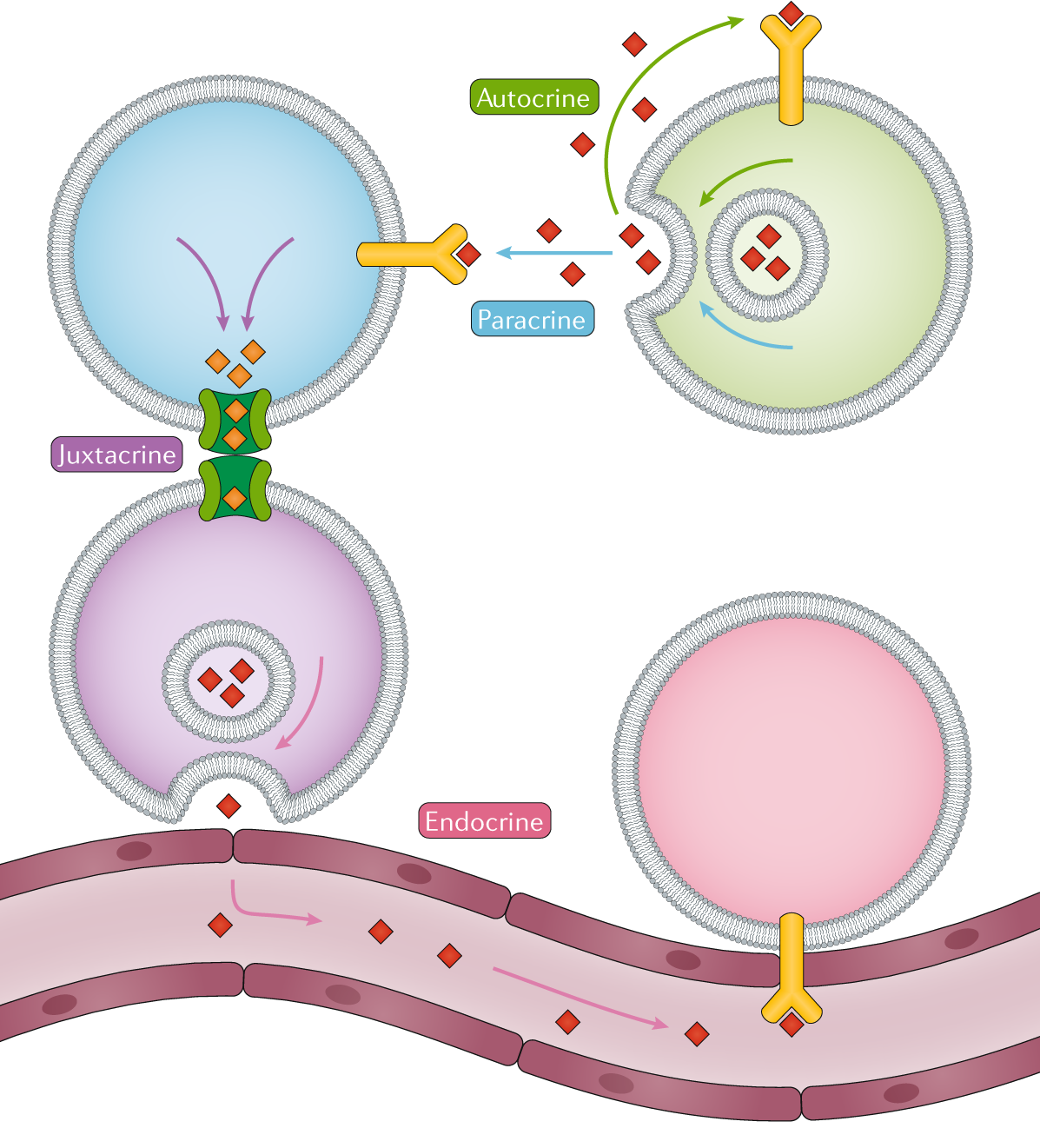
Cell-cell interactions define high-order organizations in multicellular organisms, influencing their phenotypes. In this regard, I am interested in helping to understand how CCIs and phenotype are connected, in both a cellular and molecular level.
Cellular metabolism
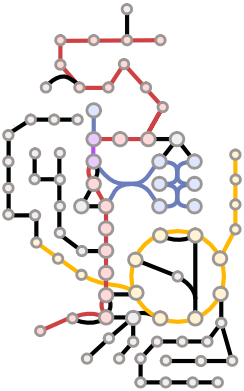
Cellular metabolism orchestrates the supply of resources and energy needed for the diversity of cellular functions. I aim to quantify metabolic activities within individual cells by using metabolic modeling and machine learning to understand physiological functions and dysregulation during aging and diseases.
Publications
Featured publications, presentations and talks that are available online. A full list of publications is available in Google Scholar.
Media & Press - Why AI Companies Are Racing to Build a Virtual Human Cell
![]()
Research Article - Atlas-scale metabolic activities inferred from single-cell and spatial transcriptomics
![]()
Review - The diversification of methods for studying cell–cell interactions and communication
![]()
Research Article - Inferring a spatial code of cell-cell interactions across a whole animal body
![]()
Research Article - Context-aware deconvolution of cell-cell communication with Tensor-cell2cell
![]()
Talk - Tensor-cell2cell: Desde la transcriptómica a entender cómo el contexto biológico afecta la comunicación intercelular
![]()
Talk - Computational resources for studying cell-cell communication
![]()
Review - Deciphering cell-cell interactions and communication from gene expression
![]()
Research Article - Understanding the impact of the cofactor swapping of isocitrate dehydrogenase over the growth phenotype of Escherichia coli on acetate by using constraint-based modeling
![]()
Computational suite for studying cell-cell interactions and communication

Computational suite implementing different methods to study cell-cell interactions from expression levels of genes/proteins
Docker container for single-cell analyses

Metabolic activity from single‑cell and spatial transcriptomics with scCellFie

scCellFie infers metabolic activities from single-cell and spatial transcriptomics and offers a variety of downstream analyses.
Utility for facilitating the use of COBRApy
Research Training
Postdoctoral Fellow
Wellcome Sanger InstitutePostdoctoral fellow at the Vento Lab and part-time participation at the Lotfollahi Lab. Studying the placental development and the effect of menopause on homeostasis and the dynamic resilience of women.
November 2023 - Present
Graduate Researcher
University of California, San DiegoPhD Student in Bioinformatics & Systems Biology. Worked on inferring cell-cell interactions from gene expression at the Lewis Lab. First-year lab rotations were about:
- Cell-cell interactions in the whole body of C. elegans (Dr. Nathan E. Lewis)
- Machine learning for simulating protein expression from ME-models (Dr. Bernhard Palsson)
- Relationship between microbiome composition and chronic diseases (Dr. Rob Knight)
September 2018 - July 2023
Undergraduate Researcher
Universidad de ChileWorked on my thesis project for obtaining the degree of Engineer in Molecular Biotechnology. Research conducted at the Laboratory of Biochemistry and Molecular Biology at College of Sciences. Advisor: Dr. Ricardo Cabrera Paucar. Main points of the research include:
- Used Datsenko & Wanner method to construct E. coli strains. Verification by sequencing the modified region.
- Determined physiological parameters (growth rate, carbon source uptake rate and biomass yield) of different strains. Analyzed the concentration of carbon source by HPLC.
- Simulated the metabolic behavior of strains. Used Flux Balance Analysis to explore the impact of mutations.
September 2013 – February 2015
Volunteer Undergraduate Researcher
Universidad de ChileFlux Balance Analysis of the Escherichia coli icd-NAD strain. College of Sciences. Advisor: Dr. Ricardo Cabrera Paucar. Main points include:
- Modeled the metabolic phenotype of an E. coli strain in different scenarios by using COBRA and Flux Balance Analysis.
- Analyzed data after simulations on MATLAB and created “Flux-Array” on Python to visualize differences between two flux distributions.
March 2013 – July 2013
Volunteer Undergraduate Researcher
Universidad de ChileAnalysis of Humoral and Cell-Mediated immune response in blood of patients with malignant melanoma. College of Medicine. Advisor: Dr. Flavio Salazar Onfray. Main points include:
- Performed ELISA to quantify antigens, western blot to analyze humoral response of patients and flow cytometry to study cell populations in blood.
- Maintained and cultured melanoma cell lines.
August 2011 – March 2012
Volunteer Undergraduate Researcher
Universidad de ChileNicotinic profile of motor activity in adult Zebrafish. College of Sciences. Advisor: Dr. Patricio Iturriaga Vasquez. Main points include:
- Maintenance and care of Zebrafish (Danio rerio).
- Analyzed the behavior of Zebrafish in a trapezoid model of tank after immersing them in a beaker with different concentrations of nicotine and other drugs.
- Tracked the movement of Zebrafish in the tank by using DanioTrack.
May 2010 – December 2010
Honors & Awards
2024 Marie Skłodowska-Curie Actions Fellowship
European CommissionPostdoctoral Fellowship
2025 - 2027
2023-24 Jean Fort Dissertation Prize
University of California, San DiegoThe prize is awarded annually to a Ph.D. recipient of unusual intellectual breadth whose doctoral research has met the highest standards of academic excellence and may make a significant contribution on an issue of humanitarian or public concern.
2024
Siebel Scholar, Class of 2023
Siebel Scholars FoundationAwarded annually for academic excellence and demonstrated leadership to over 90 top students from the world’s leading graduate schools.
2022 - 2023
Academic Hardware Grant
NVIDIAAwarded a NVIDIA RTX A6000 for developing Tensor-cell2cell
2022
Fellowship by the Government of Chile
ANID - Becas ChilePredoctoral fellowship
2018 - 2022
Fulbright Fellowship
Fullbright CommissionPredoctoral Fellowship
2018 - 2022
Winning Talk
ANEIB ChileBest oral presentation at the II National Congress of Biotechnology Students.
2013
Skills
- Python
- R
- git
- linux
- bash
- MySQL
- SQLite
- Arduino
- Raspberry Pi
- Fútbol
- Magic the Gathering
- Electric Guitar
- Photography